2. Loading Data¶
In a visualization pipeline, data sources bring data into the system for
processing and visualization. Sources, such as the Sphere source
(accessible from the Sources menu in paraview),
programmatically create datasets for
processing. Another type of data sources are readers. Readers can read data
written out in disk files or other databases and bring it into ParaView for
processing. ParaView includes readers that can read several of the
commonly used scientific data formats. It’s also possible to write plugins that
add support for new or proprietary file formats.
ParaView provides several sample datasets for you to get started. You can download an archive with several types of data files from the download page at https://www.paraview.org/download under the Data section.
2.1. Opening data files in paraview¶
To open a data file in paraview, you use the Open File dialog.
This dialog can be accessed from the File > Open menu or by using the
![]() button in the
button in the Main Controls toolbar. You can
also use the keyboard shortcut CTRL + O (or ⌘ + O) to open this dialog.
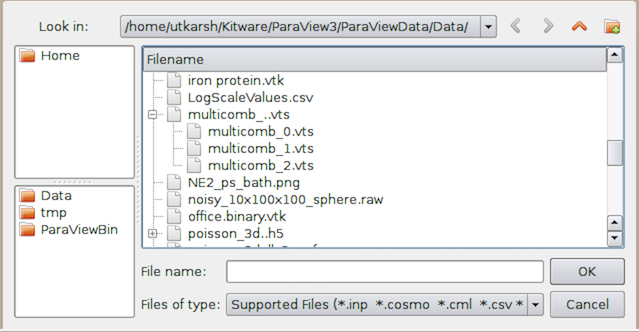
Fig. 2.1 Open File dialog in paraview for opening data (and other) files.¶
The Open File dialog
allows you to browse the file system on the data
processing nodes. This will become clear when we look at using ParaView for
remote visualization. While several of the UI elements in this dialog are
obvious such as navigating up the current directory, creating a new
directory, and navigating back and forth between directories, which can all be done with the
standard system shortcuts like CTRL + N (or ⌘ + N) to create a directory or
Alt + ↑ to go to the parent directory, there are a few things to note.
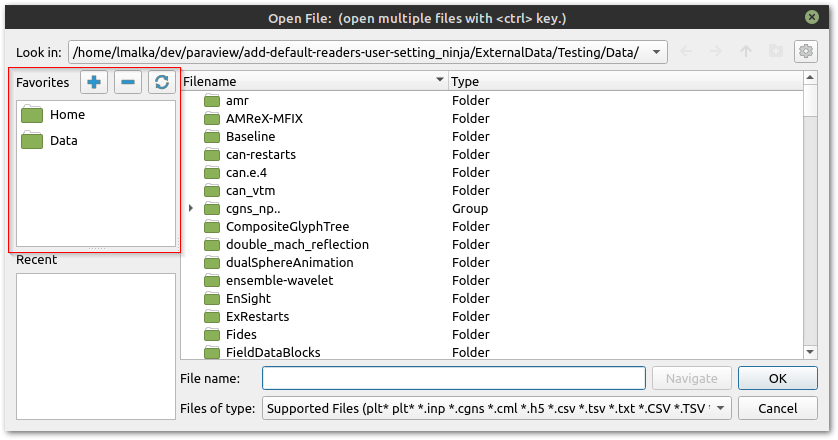
The
Favoritespane shows some platform-specific common locations such as the home directory and desktop. The favorites directories can be customized from the buttons above it, which respectively adds the current directory to the favorites, removes the current directory from the favorites, and resets all the favorites to the system default. Another way to change which directories are in the favorites is to use the right-click context menus. Right-clicking any directory in the main list will display a menu with the option to add it to the favorites. A right-click on a favorite in the favorites list displays the option to remove it.The
Recent Directoriespane shows a few of the most recently used directories.
You can browse to the directory containing your datasets and either select the
file and hit Ok or simply double click on the file to open it. You can also
select multiple files using the CTRL (or ⌘) key. This will open
each of the selected files separately.
Right-clicking the files list will display a few options, depending on what was right-clicked.
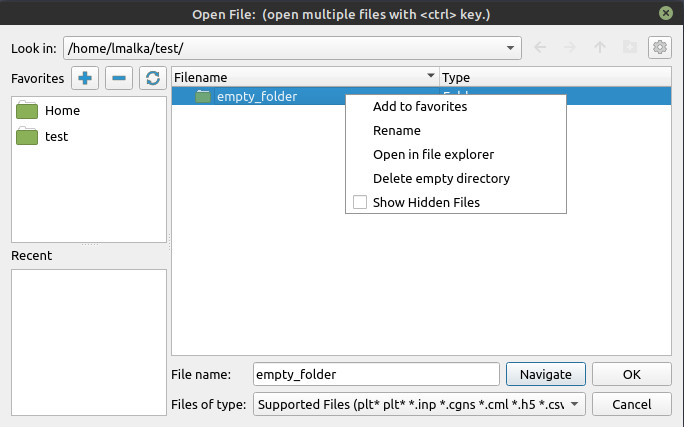
The
Show Hidden Filesis always visible, and can be checked to display the hidden files and directories.The
Open in file exploreris also always present and either opens in the system file explorer the selected directory if one was right-clicked, or opens the current directory if a file or nothing was selected.Selecting a file or a directory adds the
Renameoption.Selecting a folder adds the
Add to favoritesoption which adds the selected directory to theFavoritespane on the left.Selecting an empty directory provides the
Delete empty directoryoption.
When a file is opened, paraview will create a reader instance of
the type suitable for the selected file based on its extension. The reader will
simply be another pipeline module, similar to the source we created in
Section 1. From this point forward, the workflow will be
the same as we discussed in Section 1.4.2 :
You adjust the reader properties, if needed, and hit Apply .
paraview will then read the data from the file and render it in the
view.
If you selected multiple files using the CTRL (or ⌘) key,
paraview will create multiple reader modules. When you hit
Apply , all of the readers will be executed, and their data will be shown in the view.
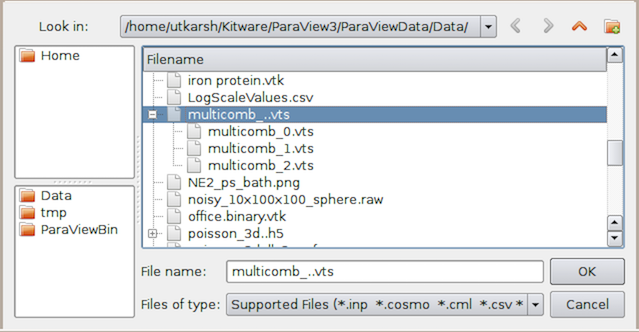
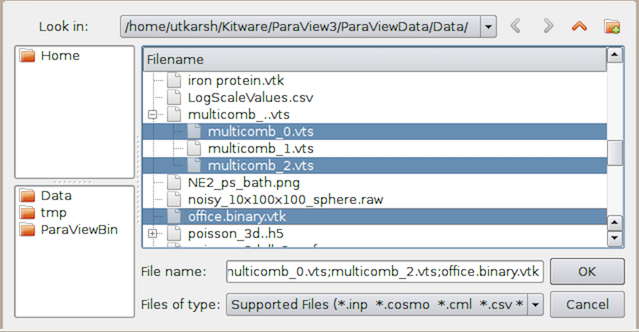
Fig. 2.2 The Open File dialog can be used to select a
temporal file series (top) or select multiple files to open separately (bottom).¶
Did you know?
This ability to hit the Apply button once to accept changes on multiple
readers applies to other pipeline modules, including sources and filters.
In general, you can change properties for multiple modules in a pipeline, and
hit Apply to accept all of the changes at once. It is possible to
override this behavior from the Settings dialog.
2.1.1. Dealing with unknown extensions¶
On occasion, you will run into situations where a file has an unusual name and,
despite the fact that ParaView supports reading the file format,
paraview does not recognize the file because its extension does not
match the expected extension. In this case, paraview will pop
up the Open Data With... dialog, which lists several readers
(Fig. 2.3).
You can then pick the reader for the correct file format from this list
and continue. If you picked an incorrect reader, however, you’ll get error
messages either when the reader module is instantiated or after you hit
Apply . In either case, you can simply Delete the reader module and try
opening the file again, this time choosing a different reader.
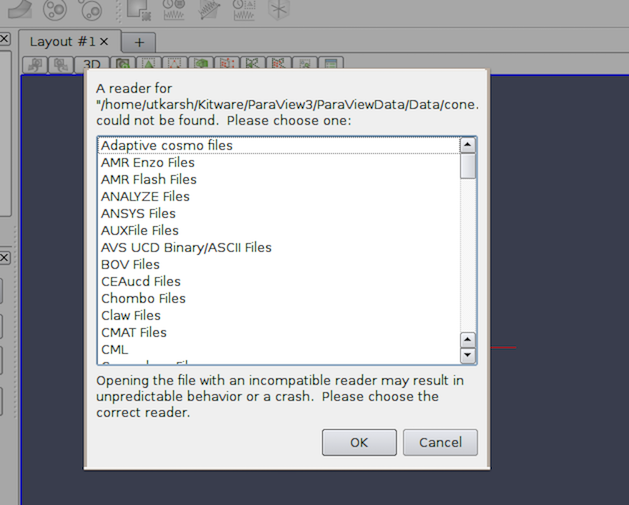
Fig. 2.3 Open Data With... dialog shown to manually choose the reader to use for a file with an unknown extension.}¶
Error messages in paraview are shown in the Output Messages
window (Fig. 2.4).
It is accessible from the View > Output Messages menu.
Whenever there’s a new error message, paraview will
automatically pop open this window and raise it to the top. This window can
be attached, or docked, in the main window so that it is visible with the
other user interface elements without covering them up.
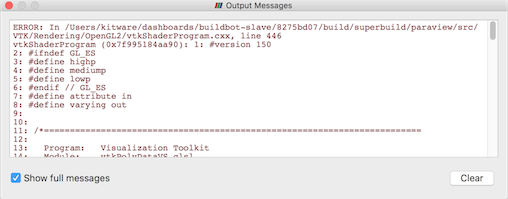
Fig. 2.4 The Output Messages window is used to show errors, warnings, and other messages raised by the application.¶
2.1.2. Handling temporal file series¶
Most datasets produced by scientific simulation runs are temporal in nature. File formats differ in how this information is saved in the file. While several file formats support saving multiple timesteps in the same file, others save them out as a sequence of files, known as a file series
The Open File dialog automatically detects file series and shows them as a
grouped element, as shown in Fig. 2.2. To load the file series,
simply select the group, and hit Ok . You can also open a single file in the
series as a regular file. To do so, open the file group and select the file
you want to open.
paraview automatically detects several of the commonly-known file
naming patterns used for indicating a file series. These include:
|
|
|
|
|
|
|
|
where foo could be any filename, N is a numeral sequence (with any number of leading zeros), and vtk could be any extension.
2.1.3. Dealing with time¶
When you open a dataset with time, either as a file series or in a file format
that natively supports time, paraview will automatically setup an
animation for you so that you can play through each of the time steps in the
dataset by using the  button
on the
button
on the VCR Controls toolbar (Fig. 2.5).
You can change or modify this animation and
further customize it, as discussed in Chapter Section 7.

Fig. 2.5 VCR Controls toolbar for interacting with an animation.¶
2.1.4. Reopening previously opened files¶
paraview remembers most recently opened files (or file series).
Simply use the File > Recent Files menu. paraview also
remembers the reader type selected for files with unknown extensions or for occasions when
multiple reader choices were available.
2.1.5. Opening files using command line options¶
paraview provides a command line option that can be used to open
datasets on startup.
> paraview --data=.../ParaViewData/Data/can.ex2
This is equivalent to opening a can.ex2 data file from the Open File dialog.
The same set of follow-up actions happen. For example, paraview will try to
locate a reader for the file, create that reader, and wait for you to hit
Apply .
To open a file series, simply replace the numbers in the file name sequence by
a . For example, to open a file series named my0.vtk, my1.vtk … myN.vtk, use my..vtk.
> paraview --data=.../ParaViewData/Data/my..vtk
2.1.6. Common properties for readers¶
ParaView uses different reader implementations for different file formats. Each of these have different properties available to you to customize how the data is read and can vary greatly depending on the capabilities of the file format itself or the particular reader implementation. Let’s look at some of the properties commonly available in readers.
2.1.6.1. Selecting data arrays¶
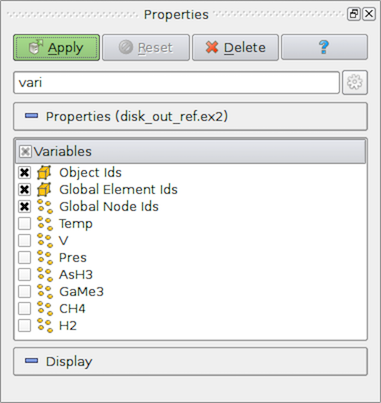
Fig. 2.6 Array selection widget for selecting array to load from a data file.¶
One of the most common properties on readers is one that allows you to select the data arrays (cell centered, point centered, or otherwise, if applicable) to be loaded. Often times, loading only the data arrays you know you are going to use in the visualization will save memory, as well as processing time, since the reader does not have to read in those data arrays, and the filters in the pipeline do not have to process them.
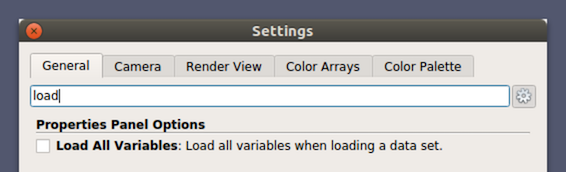
Did you know?
You can change paraview’s default behavior to load all available data arrays by
selecting the Load All Variables checkbox under
Settings/Properties Panel Options/Advanced .
The user interface for selecting the arrays to load is simply a list with the names of the
arrays and a checkbox indicating whether that array is to be loaded or not
(Fig. 2.6). Icons, such as  and
and  are often
used in this widget to give you an indication of whether the array is
cell-centered or point-centered, respectively.
are often
used in this widget to give you an indication of whether the array is
cell-centered or point-centered, respectively.
If you initially de-select an array, but then as you’re setting up your visualization pipeline
realize that you need that data array, you can always go back to the
Properties page for the reader by making the reader active in the
Pipeline Browser and then changing the array selection. ParaView will
automatically re-execute any processing pipeline set up on the reader with this
new data array.
Common Errors
Remember to hit Apply (or use Auto Apply ) after changing the array
selection for the change to take effect.
Sometimes the list of data arrays can get quite large, and it can
become cumbersome to find the array for which you are looking. To help
with such situations, paraview provides a mechanism to search lists. Click inside the
widget to make it get the focus. Then type CTRL + F (or
⌘ + F) to get a search widget. Now you can type in the text to
search. Matching rows will be highlighted (Fig. 2.7).
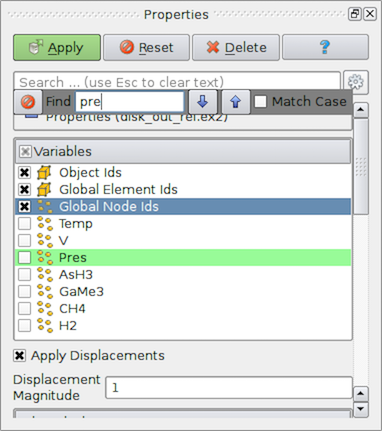
Fig. 2.7 To search through large lists in paraview, you can use CTRL + F.¶
Did you know?
The ability to search for items in an array selection widget also applies to
other list and tree widgets in the paraview UI. Whenever
you see a widget with a large number of entries in a list, table, or tree fashion,
try using CTRL + F (or ⌘ + F).
2.2. Opening data files in pvpython¶
To open data files using the scripting interface, ParaView provides the
OpenDataFile function.
>>> reader = OpenDataFile(".../ParaViewData/Data/can.ex2")
>>> if reader:
... print("Success")
... else:
... print("Failed")
...
OpenDataFile will try to determine an appropriate reader based on the file
extension, just like paraview. If no reader is determined,
None is returned. If multiple readers can open the
file, however, OpenDataFile simply picks the first reader. If you
explicitly want to create a specific reader, you can always create the reader by
its name, similar to other sources and filters.
>>> reader = ExodusIIReader(FileName=".../ParaViewData/Data/can.ex2")
To find out information about the reader created and the properties available on it,
you can use the help function.
>>> reader = ExodusIIReader(FileName=".../ParaViewData/Data/can.ex2")
>>> help(reader)
Help on ExodusIIReader in module paraview.servermanager object:
class ExodusIIReader(ExodusIIReaderProxy)
| The Exodus reader loads
| Exodus II files and produces an unstructured grid output.
| The default file extensions are .g, .e, .ex2, .ex2v2,
| .exo, .gen, .exoII, .exii, .0, .00, .000, and .0000. The
| file format is described fully at:
| http://endo.sandia.gov/SEACAS/Documentation/exodusII.pdf.
| ...
|
| -----------------------------------------------------------------
| Data descriptors defined here:
|
| AnimateVibrations
| If this flag is on and HasModeShapes is also on, then
| this reader will report a continuous time range [0,1] and
| animate the displacements in a periodic sinusoid. If this
| flag is off and HasModeShapes is on, this reader ignores
| time. This flag has no effect if HasModeShapes is off.
|
| ApplyDisplacements
| Geometric locations can include displacements. When this
| option is on, the nodal positions are 'displaced' by the
| standard exodus displacement vector. If displacements are
| turned 'off', the user can explicitly add them by applying
| a warp filter.
| ...
Did you know?
The help function can be used to get information about properties available on
any source or filter instance. It not only lists the properties, but also
provides information about how they affect the pipeline module. help can
also be used on functions. For example:
>>> help(OpenDataFile)
Help on function OpenDataFile in module paraview.simple:
OpenDataFile(filename, \*\*extraArgs)
Creates a reader to read the given file, if possible.
This uses extension matching to determine the best reader
possible. If a reader cannot be identified, then this
returns None.
2.2.1. Handling temporal file series¶
Unlike paraview, pvpython does not automatically
detect and load file series . You have to explicitly list the filenames in the
series and pass that to the OpenDataFile call.
# Create a list with the names of all the files in the file series in
# correct order.
>>> files = [".../Data/multicomb_0.vts",
".../Data/multicomb_1.vts",
".../Data/multicomb_2.vts"]
>>> reader = OpenDataFile(files)
2.2.2. Dealing with time¶
Similar to paraview, if you open a time series or a file with
multiple timesteps, pvpython will automatically set up an animation
for you to play through the timesteps.
>>> files = [".../Data/multicomb_0.vts",
".../Data/multicomb_1.vts",
".../Data/multicomb_2.vts"]
>>> reader = OpenDataFile(files)
>>> Show()
>>> Render()
# Get access to the animation scene.
>>> scene = GetAnimationScene()
# Now you use the API on the scene when doing things such as playing
# the animation, stepping through it, etc.
# This will simply play through the animation once and stop. Watch
# the rendered view after you hit `Enter.'
>>> scene.Play()
2.2.3. Common properties on readers¶
2.2.3.1. Selecting data arrays¶
For those properties on readers that allow you to control what to read in from
the file such as point data arrays, cell data arrays, or data blocks,
paraview uses a selection widget, as seen in
Section 2.1.6.1. Likewise, pvpython provides
an API that allows you to determine the available options and then
select/deselect them.
The name of the property that allows you to make such selections depends on the
reader itself. When in doubt, use the tracing capabilities in
paraview (Section 1.6.2) to figure it out. You can also use
help (Section 2.2).
ExodusIIReader has a PointVariables property that can be used to
select the point data arrays to load. Let’s use this as an example.
# Open an ExodusII data file.
>>> reader = OpenDataFile(".../Data/can.ex2")
# Alternatively, you can explicitly create the reader instance as:
>>> reader = ExodusIIReader(FileName = ".../Data/can.ex2")
# To query/print the current status for `PointVariables' property,
# we do what we would have done for any other property:
>>> print(GetProperty("PointVariables"))
['DISPL', 'VEL', 'ACCL']
# An alternative way of doing the same is as follows:
>>> print(reader.PointVariables)
['DISPL', 'VEL', 'ACCL']
# To set the property, simply set it to list containing the names to
# enable, e.g., if we want to read only the 'DISPL' array, we do
# the following:
>>> SetProperties(PointVariables=['DISPL'])
# Or using the alternative way for doing the same:
>>> reader.PointVariables = ['DISPL']
# Now, the new value for PointVariables is:
>>> print(reader.PointVariables)
['DISPL']
# To determine the array available, use:
>>> print(reader.PointVariables.Available)
['DISPL', 'VEL', 'ACCL']
# These are the arrays available in the file.
Changing PointVariables only changes the value on the property. The reader
does not re-execute until a re-execution is requested either by calling
Render or by explicitly updating the pipeline using UpdatePipeline .
>>> reader.PointVariables = ['DISPL', 'VEL', 'ACCL']
# Assuming that the reader is indeed the active source, let's update
# the pipeline:
>>> UpdatePipeline()
# Or you can use the following form if you're unsure of the active
# source or just do not want to worry about it.
>>> UpdatePipeline(proxy=reader)
# Print the list of point arrays read in.
>>> print(reader.PointData[:])
[Array: ACCL, Array: DISPL, Array: GlobalNodeId, Array: PedigreeNodeId, Array: VEL]
# Change the selection.
>>> reader.PointVariables = ['DISPL']
# Print the list of point arrays read in, nothing changes!
>>> print(reader.PointData[:])
[Array: ACCL, Array: DISPL, Array: GlobalNodeId, Array: PedigreeNodeId, Array: VEL]
# Update the pipeline.
>>> UpdatePipeline()
# Now the arrays read in has indeed changed as we expected.
>>> print(reader.PointData[:])
[Array: DISPL, Array: GlobalNodeId, Array: PedigreeNodeId]
We will cover the reader.PointData API in more details in Section 3.3.
2.3. Reloading files¶
While ParaView is often used after the simulation has generated all the data,
it is not uncommon to use ParaView to inspect data files as they are being
written out by the simulation. In such cases, the simulation may either be
modifying existing file(s) with new timesteps or creating
new files for each timestep. In such cases, you may want to refresh
ParaView to make it aware of the changes. In paraview, this can be done using
Reload Files .
When the reader is active, you can use the File > Reload Files menu to
request the reader to refresh. paraview will prompt you to choose whether to
reload the existing file(s) or look for new files in the file series, as shown in
Fig. 2.8. Click on Reload existing file(s) , to force
the reader to re-read the files already opened. This is useful in cases
where the simulation may have modified existing file(s).
Use Find new files to make the reader aware of any new files in the
file series.
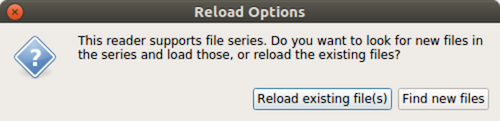
Fig. 2.8 The Reload Options dialog allows you to choose how to refresh the reader.¶
Similar to paraview, in pvpython, you use ReloadFiles to reload existing files, and
ExtendFilesSeries to look for new files in a file series.
# For file being modified in place per timestep
>>> reader = OpenDataFile(file)
...
>>> ReloadFiles(reader)
# For files being generated per timestep
>>> reader = OpenDataFile(file)
...
>>> ExtendFilesSeries(reader)